Fluorescent RNA Technology
RNA is an emerging, rapidly growing research field in biological science and medicine. In addition to the well known functions of mRNAs, rRNAs, tRNAs in the central dogma of molecule biology, recent studies reveal the identity and functions of vast noncoding RNAs (ncRNAs) that play an important role in diverse biological processes, which are reshaping the prior conceptions about RNA functions. In living cells, RNAs exhibit complex dynamics including expression, degradation, translocation, splicing and various chemical modifications.
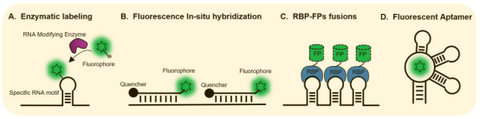
Methodology to visualize RNA such as is fluorescent in situ hybridization (FISH), enzymatic covalent labeling require cell fixation and are not suitable for live cell imaging. RNAs with engineered motifs can be tethered with fusions of fluorescent protein and specific RNA binding proteins (RBPs) e.g. MCP, PCP, λN or Cas may be used to image RNA in live cells at the single molecule level. However, the unbound MCP-FP molecules diffuse throughout the cells and generates high background fluorescence. In addition, whether such a heavy load of tethered protein affects the localization, stability and behavior of RNAs remains to be determined.
Figure 1. Technology for in situ RNA fluorescent labeling and detection.
In the history, fluorescent proteins (FPs) of different colors had revolutionized research of life sciences, which are genetically encoded labels of proteins enabling background free tracing of proteins in live cells. RNAs of interest may also be genetically labeled similarly and straightforwardly with fluorophore-binding RNA aptamers. These aptamers, termed fluorescent RNAs (FRs), shall also enable easy, robust, background free imaging of diverse RNAs in living cells.
While simple and as promising as they appear, however, the utility of FRs current available is limited. Some of the dye ligands of current FRs show significant background fluorescence in live cells and/or do not readily diffuse across plasma membranes, or the FRs. Some FRs has limited stability and brightness in live cells, or function as multimer. Analogously to FPs, ideal FRs should be monomeric, stable, bright, and multicolored, which had been challenging to achieve.
Peppers, a series of monomeric, highly bright, and stable FRs with a broad range of emission maxima spanning from cyan to red, were obtained by unique design of dendritic cell permeable dye ligand and multiple rounds of optimization. These dyes showed good membrane permeability, low cytotoxicity, and little fluorescence in solution or live cells. Compared to currently available FRs, Peppers showed an order of magnitude enhanced cellular fluorescence intensity and fluorescence turn-on ratio, one or two orders of magnitude enhanced affinity, ~20 oC increased Tm, expanded pH tolerance, and a broad spectral range available for live cell studies. For the first time, Peppers allow simple and robust imaging of mRNA and other RNA species in live cells with minimal perturbation of the target RNA’s transcription, localization, and translation. Peppers may also be used in imaging of genomic loci through CRISPR display, real-time tracking of protein-RNA tethering, and super-resolution imaging of RNA by structured illumination microscopy. Due to its high signal to background ratio, Peppers can be used in quantitative studies of RNAs in live cells, using flow cytometry or microplate reading. These Pepper FRs provide ideal tools for live imaging of cellular RNAs.
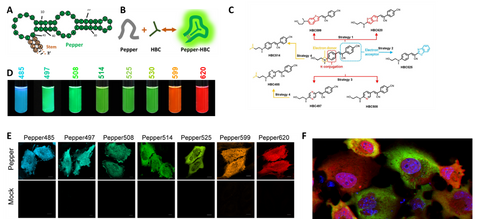
Figure 2. Pepper for background free RNA imaging and quantification in live cells. (A-C) Pepper aptamer and HBC ligands. (D) a platelet of fluorescent RNAs. (E) Background free imaging of RNAs in live cells with Pepper. (F) Multiplex imaging of RNA and its protein in live cells showed heterogenous translation in cancer cells.
Clivias is a series of small, monomeric and stable orange-to-red fluorescent RNAs with large Stokes shifts of up to 108 nm, enabling the simple and robust imaging of RNA with minimal perturbation of the target RNA’s localization and functionality. In combination with Pepper fluorescent RNAs, the Clivias enable the single-excitation two-emission dual-color imaging of cellular RNAs and genomic loci. Clivias can also be used to detect RNA–protein interactions by bioluminescent imaging both in live cells and in vivo. We believe that these large Stokes shift fluorescent RNAs will be useful tools for the tracking and quantification of multiple RNAs in diverse biological processes.
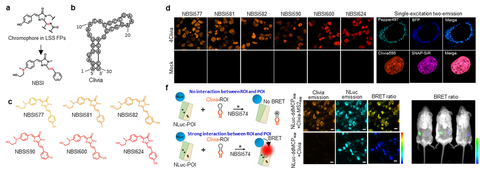
Figure 3. Cliva for background free RNA localization and interaction studies in live cells and live animal. (a-c) Clivia and its NBSI ligands. (b) Background free imaging of RNAs with Clivas. (e) Clivia and Pepper can be imaged simultaneously. (f) BRET assay of RNA-protein imaging using Nanoluc and Clivia. RNAs can be labeled and measured using BRET imaging in live animal.
We just released a highly bright green fluorescent RNA, termed Okra. Okra binds and activates the fluorophore ligand ACE to emit green fluorescence. Okra has an order of magnitude enhanced cellular brightness than currently available green FRs, allowing the robust imaging of messenger RNA in both live bacterial and mammalian cells. Due to its high brightness and photostablity, Okra is useful for time-resolved measurements of mRNA tracking to stress granules, as well as live-cell dual-color superresolution imaging of RNA in combination with Pepper620, revealing nonuniform and distinct distributions of RNA species throughout the granules. The favorable properties of Okra make it a versatile tool for the study of RNA dynamics and subcellular localization.
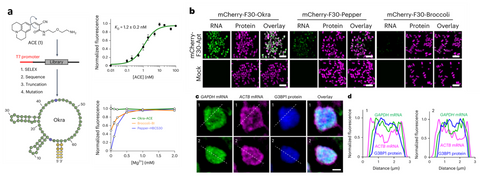
Figure 4. Okra green fluorescent RNA. (a) Okra and ACE ligand. Okra has high affinity to ACE and its fluorescence is not Mg dependant. (b) Background free imaging of RNAs with Okra, which is the brightest green FR. (c) Multicolor imaging of Okra, Pepper and fluorescent protein revealed different localization of proteins and mRNAs in the stress granule.
References
- Chen, X. et al. Visualizing RNA dynamics in live cells with bright and stable fluorescent RNAs. Nature Biotechnology. 37, 1287.doi:10.1038/s41587-019-0249-1 (2019).
- Huang, K. et al. Structure-based investigation of fluorogenic Pepper aptamer. Nature Chemical Biology 17, 1289. doi:10.1038/s41589-021-00884-6 (2021).
- Jiang, L. et al. Large Stokes shift fluorescent RNAs for dual-emission fluorescence and bioluminescence imaging in live cells. Nature Methods 2023, 20, 1563.
- Huang K et al. Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift. Nature Chemical Biology 2024, 10.1038/s41589-024-01633-1
- Zuo, F. et al. Imaging the dynamics of messenger RNA with a bright and stable green fluorescent RNA. Nature Chemical Biology 2024, 10.1038/s41589-024-01629-